QSense QCM-D technology has been used in virus research for decades. Here we show a user case example based on a peer-reviewed publication, to demonstrate how QSense technology can be used in virus research.
QSense technology is used in both fundamental and applied research. In virus-related studies, the technology is typically used to attain insights regarding virus behavior and virus interaction with the immediate surrounding.
In this user case example, which is based on the work by Parveen et. al.1, the goal was to increase the understanding of multivalent interactions and virus internalization into host cells. In this process, a single virus binds to multiple receptors in parallel, and this is an interaction which is poorly understood.
The approach in the study1, was to use a lipid-based model system to create a native-like environment for binding kinetic studies and to analyze the competition for membrane receptors. Specifically, the authors wanted to use QCM-D to understand the competition between norovirus like particles (noroVLPs) and lectin for the membrane receptors (glycosphingolipids, GSLs, embedded in the supported lipid membrane), and investigate norovirus detachment via lectin attachment. QCM-D was used to explore and answer the following questions:
To answer the questions above, QCM-D experiments were designed to mimic the scenario of interest, Fig. 1.
Model membranes (SLBs) with embedded receptors are created on top of silica sensors. The receptors are either of two different types, H type 1 or B type 1, and each type is used in three different concentrations (note that the SLB formation is not shown in the graphs, Fig. 2). Next, the model membranes are exposed to noroVLPs, then there is a buffer rinse, whereafter lectin is introduced.
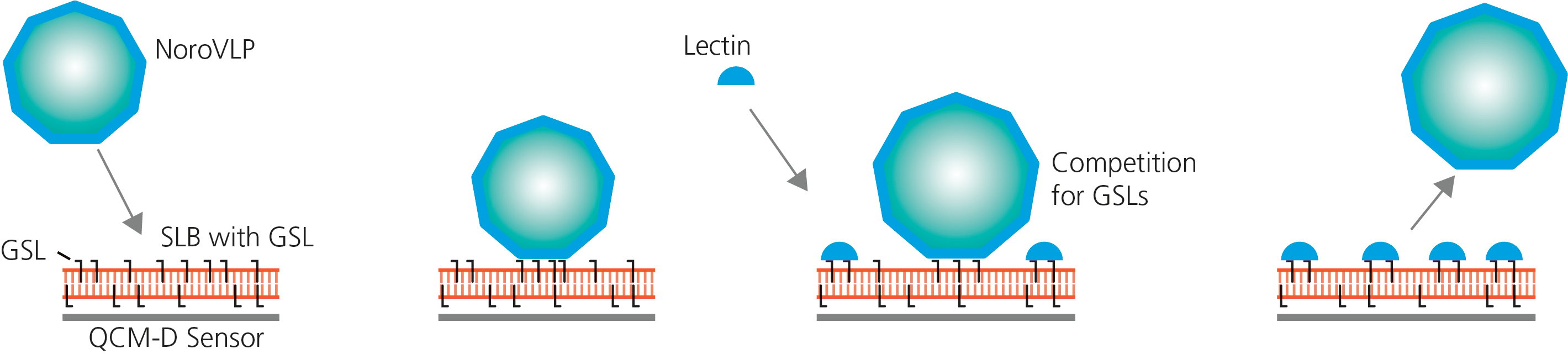
Figure 1. Schematic illustration of the lipid-based model system used to analyze competition for membrane receptors - a supported phospholipid bilayer with embedded H type 1 or B type 1 glycosphingolipids (GSLs), at different concentration, on top of an SiO2 coated QCM-D sensor. The model membrane is then exposed to noroVLPs and lectins to monitor the attachment and competition for membrane receptors.
Measurement data for the respective GSL type and concentration is shown in Fig. 2, where the left graph shows H type 1 (three concentrations) and the right graph shows H type 1 (three concentrations).
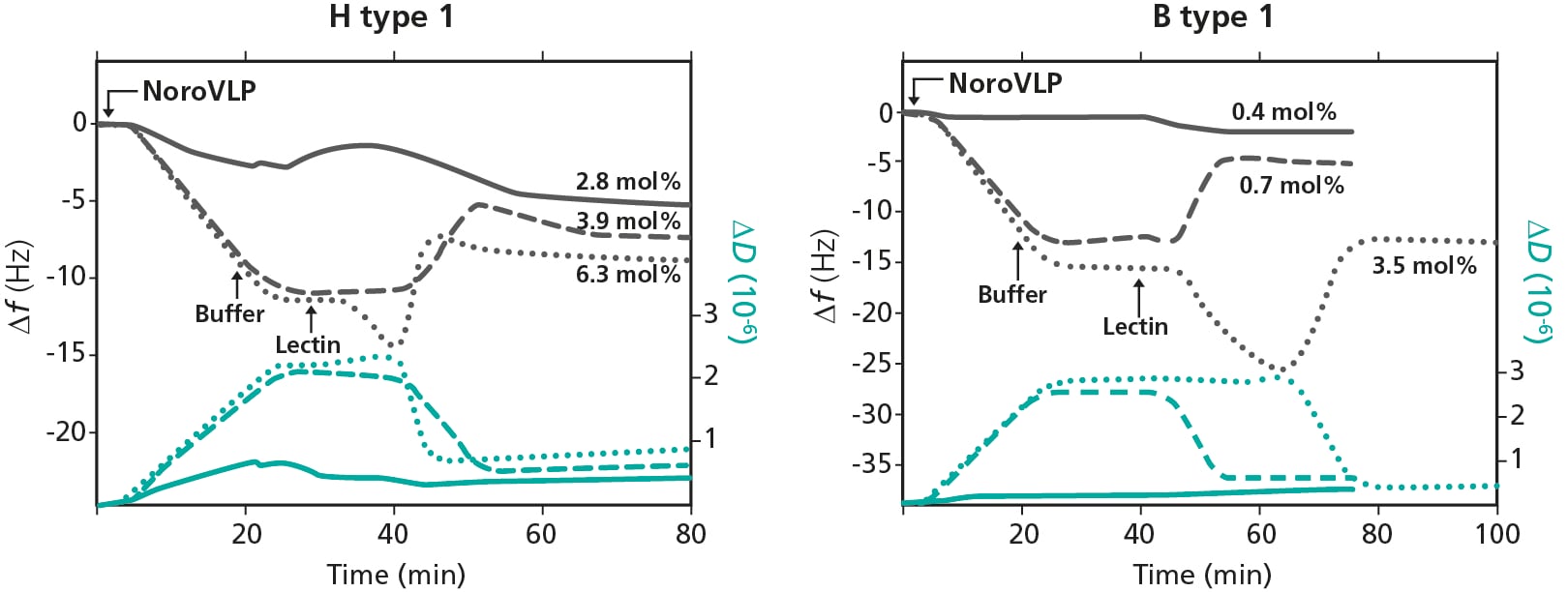
Figure 2. QCM-D measurement data from the analysis of the competition for membrane receptors. The measurements start with NoroVLP insertion, followed by a buffer rinse and then introduction of lectin. The SLB formation is not included in graphs. Left: H type 1 (three concentrations), Right: B type 1 (three concentrations).
The introduction of NoroVLPs results in a decrease in Δf, and an increase in ΔD, Fig 2. The shifts are irreversible for the medium and high GSL concentrations upon buffer rinse. These results show that the noroVLPs bind to the membrane embedded GSLs, and that the binding is irreversible upon buffer rinse with medium and high GSL concentrations. This, in turn, indicates that there is a multivalent binding of the virus at high GSL density which results in firm attachment of virus to SLB.
The Δf and ΔD shifts resulting from the injection of lectin, i.e. an increase in Δf and a decrease in ΔD, indicate that there is a release of the SLB-bound noroVLPs.
To get a more detailed understanding of the competitive behavior and to separate the detachment kinetics of noroVLPs from the lectin attachment kinetics, the authors used deconvolution of the f-trace and concluded that that lectin addition leads to a complete release of the SLB-bound noroVLPs for both GSL types.
In this study1, the goal was to explore multivalent interactions by using a lipid-based model system to create a native-like environment for binding kinetic studies. Using QCM-D, the attachment process of noroVLPs and lectin, and detachment of noroVLPs due to competition was monitored. The published work is of course much more extensive than here shown, and the analysis allowed, for example, for the quantification of:
A key takeaway emphasized by the authors is that this work demonstrates the potential of utilizing competitive binding kinetics to analyze multivalent interactions.
Download the overview on QSense analysis in virus research for additional user case examples.
Learn how QSense QCM-D reveals protein–surface interactions and adds interface-focused insight to biopharmaceutical formulation and stability work
Learn how QSense sensors enable application‑relevant biointerface interaction analysis and explore our sensor offering for different areas
Learn how QSense QCM D can be used to analyze swelling of thin films, including magnitude and dynamics.
Read about how molecule-surface interaction processes such as adsorption and desorption can be analyzed with QCM-D.
Learn best practices and step-by-step methods for accurate QCM-D coating thickness measurement on QSense sensors using QSense Omni.
Compared to QCM, QCM-D measures an additional parameter, and provides more information about the system under study.
Discover how QCM-D analysis reveals real-time etching dynamics, helping optimize cleaning processes and protect surfaces from unwanted damage.
Discover how QSense QCM-D helps tackle fouling challenges across industries
Discover how QCM-D enables real-time, label-free analysis of supported lipid membrane formation, structure, and dynamics for advanced research
